RECOMB 2025
CNGNN-DTI: Integration of Context and Neighbor Information based on Graph Neural Network for Drug-Target
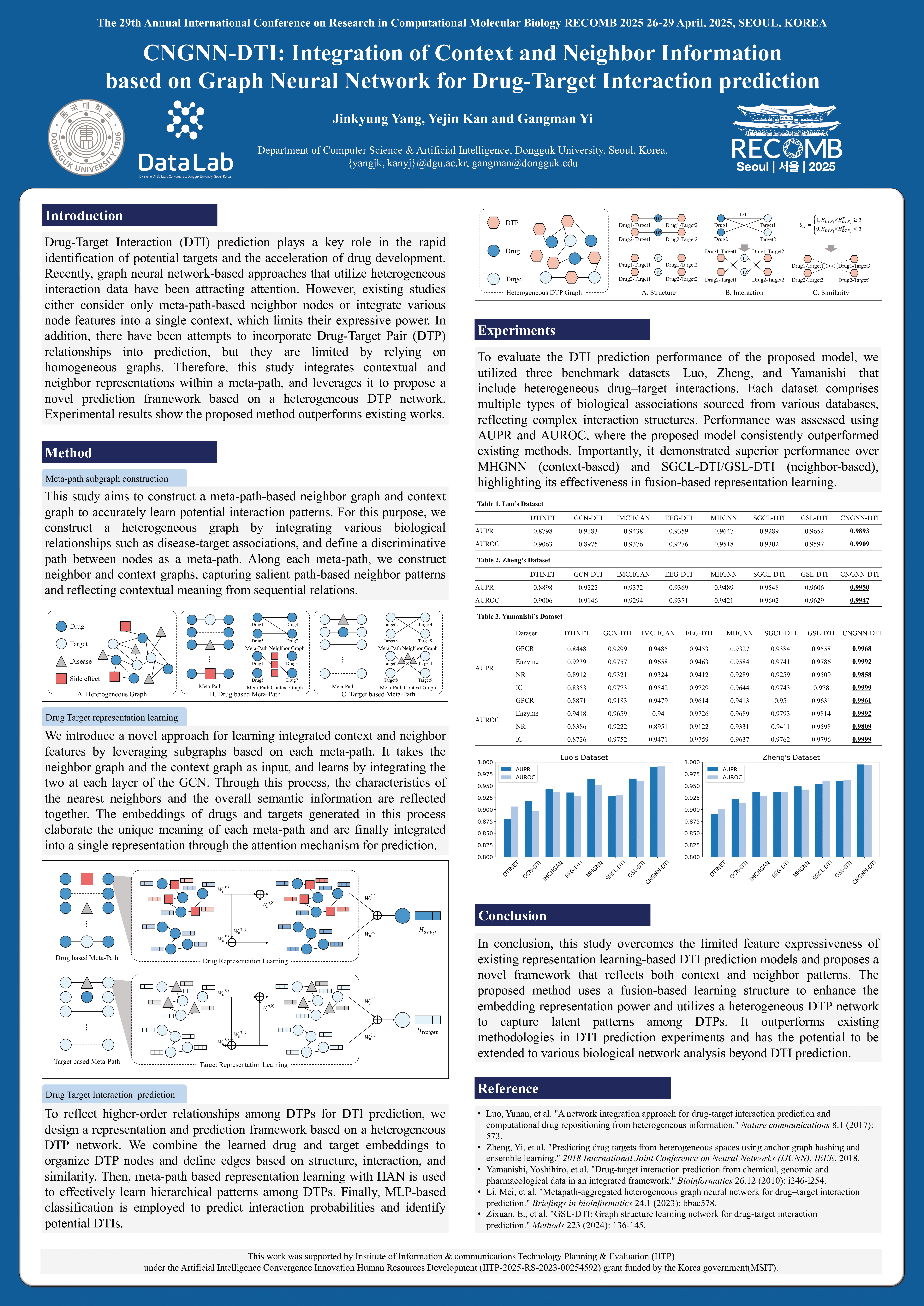
Drug-Target Interaction (DTI) prediction is essential for drug discovery and drug repurposing, and accelerates drug development. Recently, computational approaches based on graph neural networks utilizing heterogeneous biological data have been attracting attention to replace time-consuming and costly biological drug discovery approaches. In existing studies, drug and target relationships in heterogeneous biological networks are learned through various meta-paths, and Drug-Target Pair (DTP) networks are constructed to incorporate the relationships between DTPs into DTI prediction. However, context node learning is limited by considering only neighbor nodes in the meta-path, or important neighbor node information is lost by unifying instance information for context learning. In addition, existing DTP networks are built on a homogeneous graph basis, which does not fully reflect the complex correlations between DTPs. Therefore, in this study, we propose CNGNN-DTI, which can learn higher-order correlations by comprehensively learning the context and neighbor information in the meta-path and constructing a heterogeneous DTP network based on it. It independently constructs a meta-path-based neighbor graph and context graph, and learns a meaningful node vector representation through a fusion method that integrates the two information. In addition, the heterogeneous DTP network models multidimensional relationships based on structure, interaction and similarity among DTPs and integrally expresses potential interaction patterns. In conclusion, CNGNN-DTI overcomes the limitations of information loss in representation learning and captures rich correlations between DTPs to accurately capture interactions in biological systems and improve DTI prediction performance. The predicted potential DTIs can contribute to the development of new drugs in the future.