GapSense
GapSense: Similarity Estimation-Based Gap Filler with TGS-Reads for Genome Assemblies
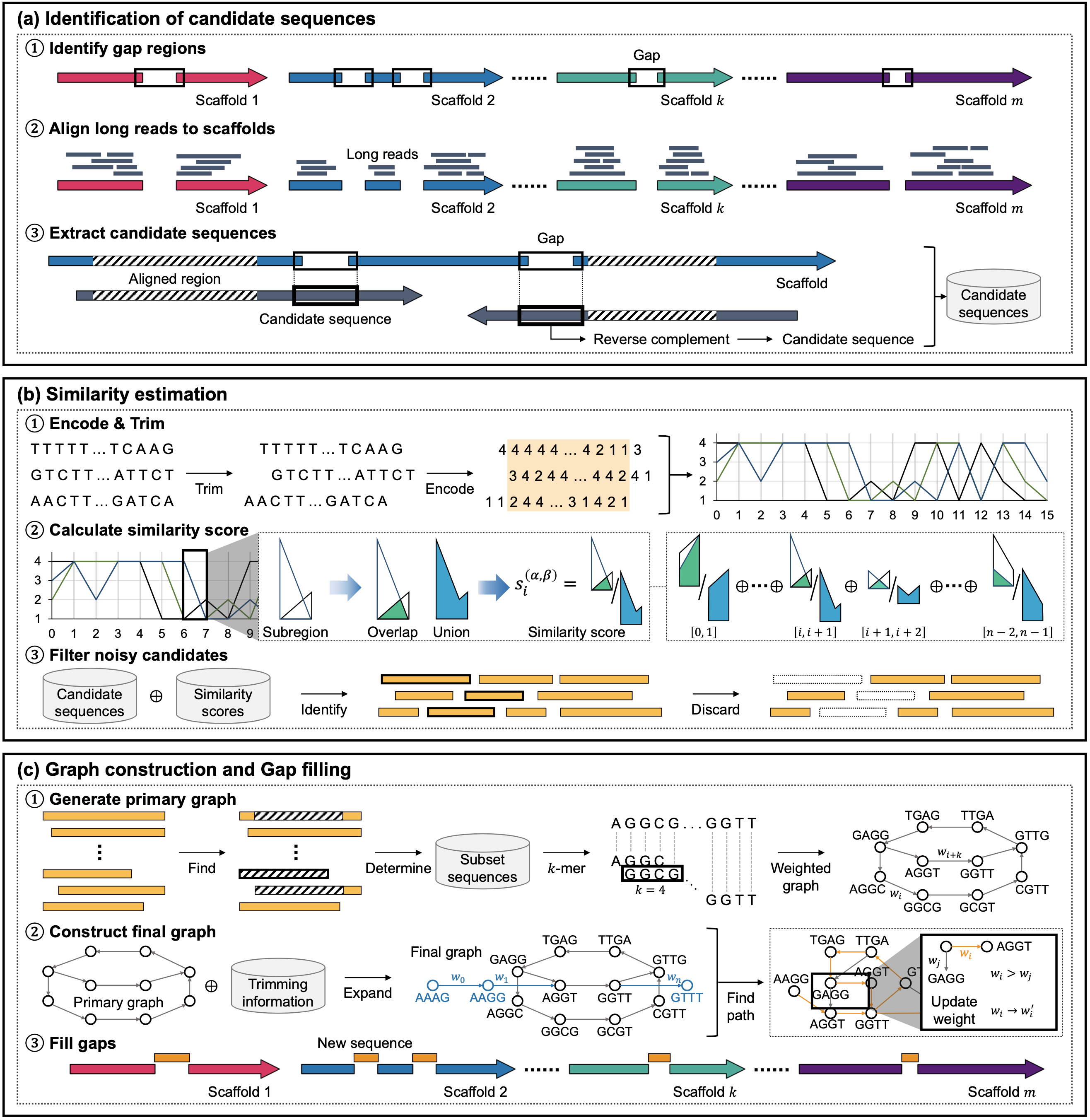
Advances in next-generation sequencing have led to an explosion in sequencing data, accelerating genome assembly research. However, draft genomes generated after scaffolding still contain unresolved gaps, often caused by repetitive regions and sequencing errors. These gaps may contain biologically meaningful sequences and thus require accurate resolution. However, existing gap-filling tools often exhibit limited reliability, especially when applied to large and complex eukaryotic genomes, due to their insufficient capacity to resolve repetitive regions or their heavy dependence on error-prone long reads. To address this challenge, we present GapSense, a robust gap-filling method that leverages similarity estimation using third-generation sequencing (TGS) reads. By quantifying pairwise similarity among candidate sequences, GapSense prioritizes informative regions and reconstructs gap sequences with higher accuracy. The proposed method introduces a novel similarity scoring mechanism that evaluates the geometric overlap of adjacent subregions to capture local structural variations and reduces noise from low-coverage and error-prone long reads. Experimental results on six representative species and three popular assemblers show that GapSense consistently outperforms existing tools in terms of gap-filling accuracy and contiguity, while maintaining low performance variability across different datasets. These findings demonstrate the effectiveness and generalizability of GapSense for accurate and scalable gap-filling.